MEASURING GENE REGULATION ACROSS LAYERS
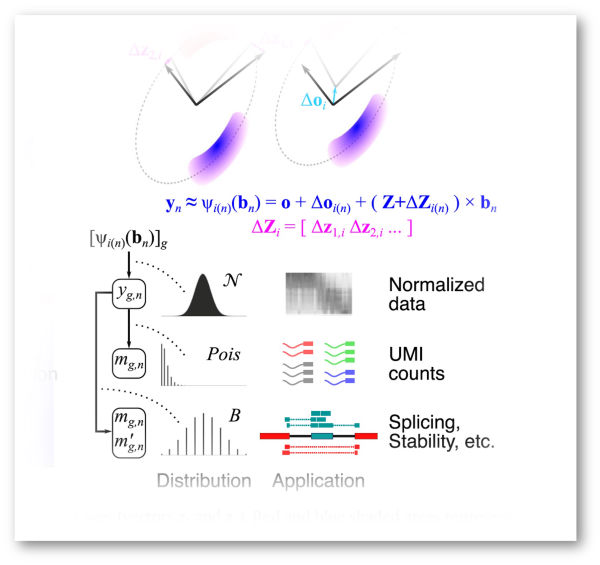
The abundance of each mRNA is shaped by the combined effects of its production, processing, and decay rates. Understanding these processes begins with accurate quantification. We combine omics technologies with novel statistical algorithms to quantify mRNA regulation—from transcription through decay and from single cells to large patient cohorts.
Example publications:
Apostolides et al., bioRxiv, 2024
Madrigal et al., Nat Commun, 2024
Farouni et al., Nat Commun, 2020
Alkallas et al., Nat Commun, 2017
SEQUENCE-TO-ACTIVITY MODELS OF GENE REGULATION
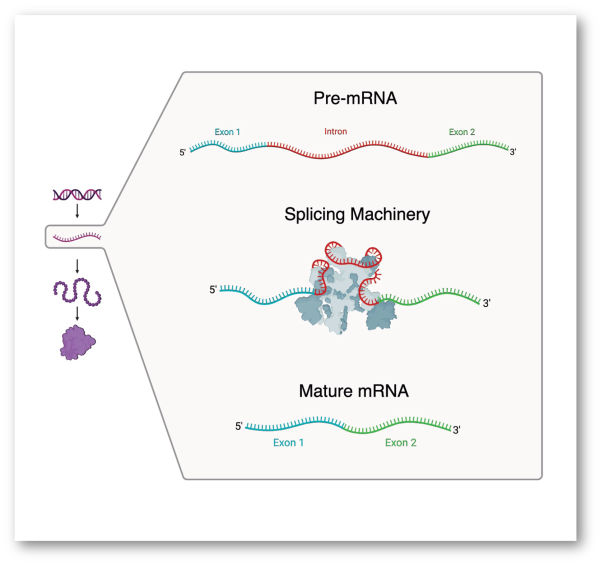
The multiple layers of mRNA regulation are governed by an extraordinarily complex network involving millions of cis-regulatory elements and trans-acting factors. We use machine learning to develop models that decode these regulatory instructions and map their interactions with transcription factors, RNA-binding proteins, and microRNAs, ultimately shaping the cell's mRNA landscape.
Example publications:
Saberi et al., bioRxiv, 2024
Corchado and Najafabadi, Genome Biol, 2022
Najafabadi et al., Nat Biotechnol, 2015
Goodarzi et al., Nature, 2012
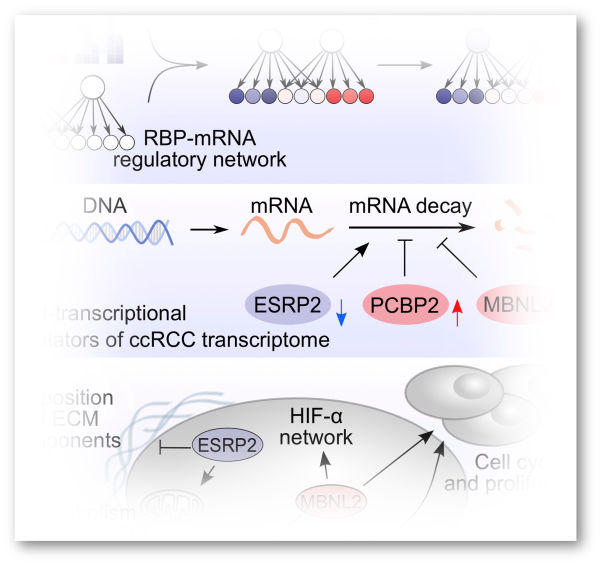
GENE REGULATORY BASIS OF CANCER
Disruptions in gene regulation drive the extensive transformation of cancer cell identity and behavior. We apply omics technologies to measure these disruptions across all layers of mRNA regulation and develop algorithms to identify the dysregulated factors that fuel cancer development and progression.
Example publications:
Perron et al., Commun Biol, 2022
Fish et al., Science, 2021
Perron et al., Cell Rep, 2018